The potential of convolutional neural networks for identifying neural states based on electrophysiological signals: experiments on synthetic and real patient data.
Deciphering our brain's signals is crucial for enhancing treatments like Deep Brain Stimulation. Machine learning is a tool capable of identifying patterns within brain data in real time. Here, we compared it to more traditional methods that focus on looking for predefined patterns. We used both simulated and real patient brain data and found that this new approach may be superior at extracting vital information from complex brain signals.
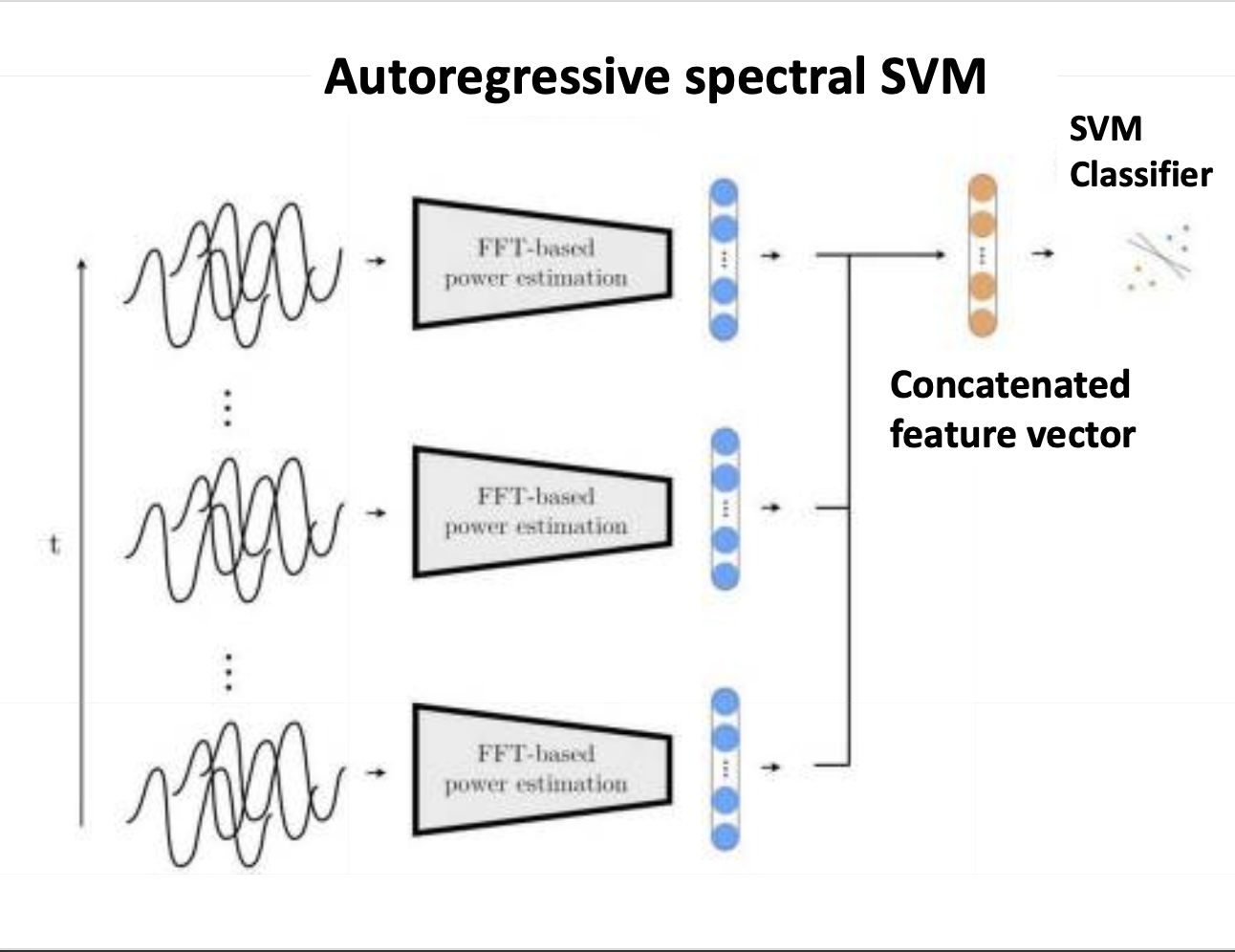
Processing incoming neural oscillatory signals in real-time and decoding from them relevant behavioral or pathological states is often required for adaptive Deep Brain Stimulation (aDBS) and other brain-computer interface (BCI) applications. Most current approaches rely on first extracting a set of predefined features, such as the power in canonical frequency bands or various time-domain features, and then training machine learning systems that use those predefined features as inputs and infer what the underlying brain state is at each given time point. However, whether this algorithmic approach is best suited to extract all available information contained within the neural waveforms remains an open question. Here, we aim to explore different algorithmic approaches in terms of their potential to yield improvements in decoding performance based on neural activity such as measured through local field potentials (LFPs) recordings or electroencephalography (EEG). In particular, we aim to explore the potential of end-to-end convolutional neural networks, and compare this approach with other machine learning methods that are based on extracting predefined feature sets. To this end, we implement and train a number of machine learning models, based either on manually constructed features or, in the case of deep learning-based models, on features directly learnt from the data. We benchmark these models on the task of identifying neural states using simulated data, which incorporates waveform features previously linked to physiological and pathological functions. We then assess the performance of these models in decoding movements based on local field potentials recorded from the motor thalamus of patients with essential tremor. Our findings, derived from both simulated and real patient data, suggest that end-to-end deep learning-based methods may surpass feature-based approaches, particularly when the relevant patterns within the waveform data are either unknown, difficult to quantify, or when there may be, from the point of view of the predefined feature extraction pipeline, unidentified features that could contribute to decoding performance. The methodologies proposed in this study might hold potential for application in adaptive deep brain stimulation (aDBS) and other brain-computer interface systems.
2023. Front Hum Neurosci, 17:1134599.
2023. Front Hum Neurosci, 17:1111590.
2021. Mov Disord, 36(4):863-873.
